Importing and visualising EEG data
Below we import and reshape EEG data from the eegkit package.
library(neurogam)
library(ggplot2)
library(eegkit)
library(dplyr)
# retrieving some EEG data
data(eegdata)
head(eegdata)
#> subject group condition trial channel time voltage
#> 1 co2a0000364 a S1 0 FP1 0 -8.921
#> 2 co2a0000364 a S1 0 FP1 1 -8.433
#> 3 co2a0000364 a S1 0 FP1 2 -2.574
#> 4 co2a0000364 a S1 0 FP1 3 5.239
#> 5 co2a0000364 a S1 0 FP1 4 11.587
#> 6 co2a0000364 a S1 0 FP1 5 14.028
# plotting the average ERP per group and channel
eegdata |>
summarise(voltage = mean(voltage), .by = c(group, channel, time) ) |>
ggplot(aes(x = time, y = voltage, colour = group) ) +
geom_line() +
facet_wrap(~channel) +
theme_bw()
# retrieving sensors x and y coordinates
data(eegcoord)
enames <- rownames(eegcoord)
# merging the data
eeg_coords <- eegcoord |>
mutate(channel = enames) |>
select(channel, xproj, yproj)
# summarising the data
eeg_data <- eegdata |>
summarise(voltage = mean(voltage), .by = c(subject, channel, time) ) |>
left_join(eeg_coords, by = "channel") |>
# converting timesteps to seconds
mutate(time = (time + 1) / 256) |>
# rounding numeric variables
mutate(across(is.numeric, ~round(.x, 4) ) ) |>
# removing NAs
na.omit() |>
# scaling the variables before fitting
mutate(
xproj = xproj / sd(xproj),
yproj = yproj / sd(yproj),
voltage = voltage / sd(voltage)
)
# sensors: one row per channel with columns xproj, yproj
sensors <- unique(eeg_data[, c("channel", "xproj", "yproj")])
# show a few rows
head(sensors)
#> channel xproj yproj
#> 1 FP1 -0.6712590 1.8012175
#> 257 FP2 0.6721339 1.7996850
#> 513 F7 -1.4744817 0.9956874
#> 769 F8 1.4718131 0.9941358
#> 1025 AF1 -0.3442040 1.4196194
#> 1281 AF2 0.3450789 1.4182019
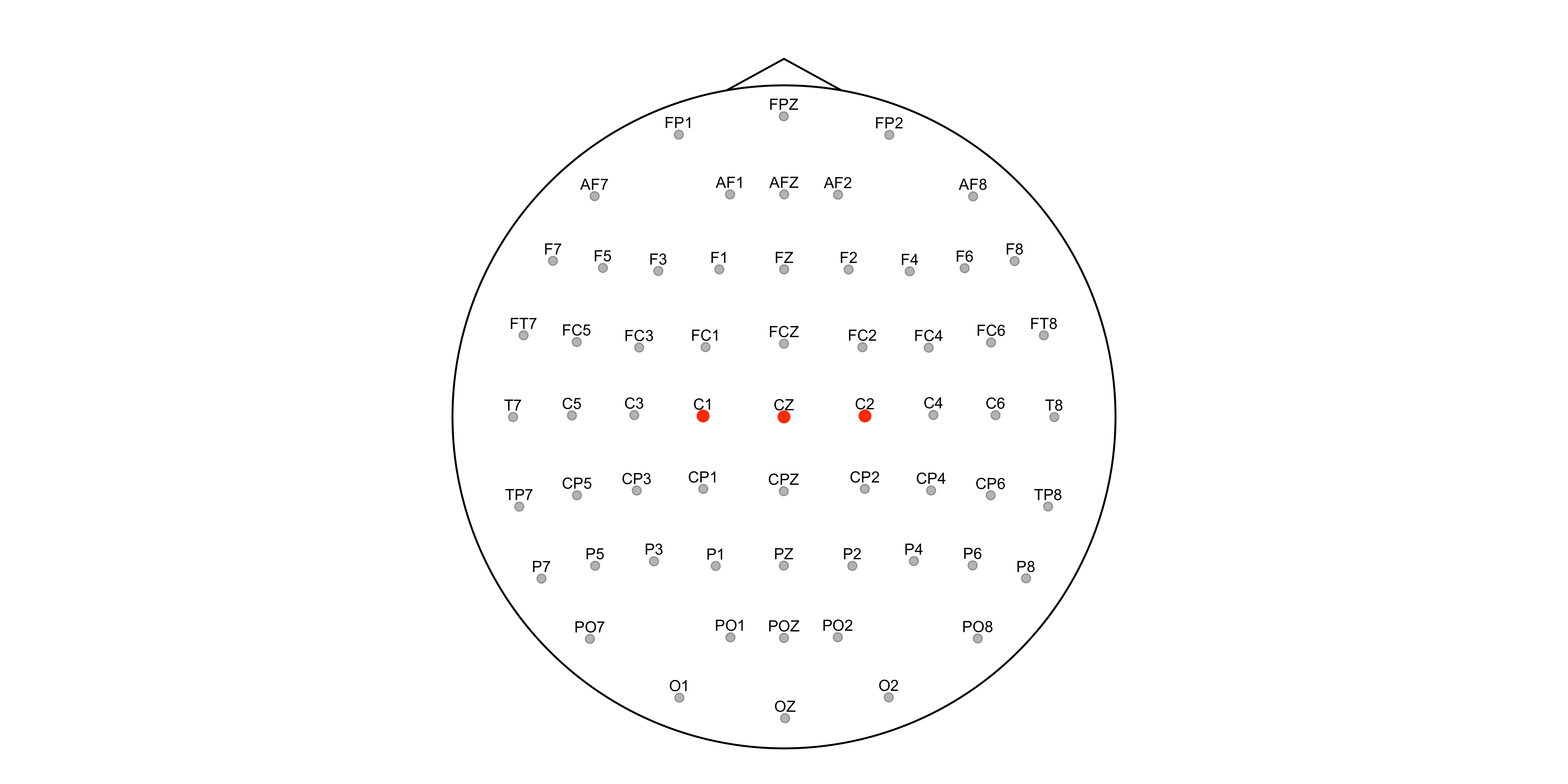
We visualise the sensors grid using the plot_sensors() function.
# plotting the sensors grid
plot_sensors(
sensors,
show_points = TRUE,
show_labels = TRUE,
label_col = "channel",
label_repel = FALSE,
highlight = c("C1", "CZ", "C2"),
label_only_highlight = FALSE,
dim_others = TRUE,
head_expand = 1.1
)
We visualise the average EEG topography through time using the plot_eeg() function.
# retrieving the first 100 unique timesteps
time_steps <- unique(eeg_data$time)[1:100]
# taking 8 equally spaced timesteps
time_steps_discrete <- st_take_n_times(time_vec = time_steps, N = 8)
# averaging EEG data per channel and timestep
eeg_data_summary <- eeg_data %>%
summarise(voltage = mean(voltage), .by = c(channel, time, xproj, yproj) ) |>
dplyr::filter(time %in% time_steps_discrete)
# topoplot of summary data
plot_eeg(
x = eeg_data_summary,
type = "topo",
sensors = sensors,
times = time_steps_discrete,
grid_res = 200,
value_col = "voltage",
facet_nrow = 2,
# fill_limits = c(-2, 2)
fill_limits = "global_quantile"
)
Spatio-temporal modelling with BGAMs
In progress.